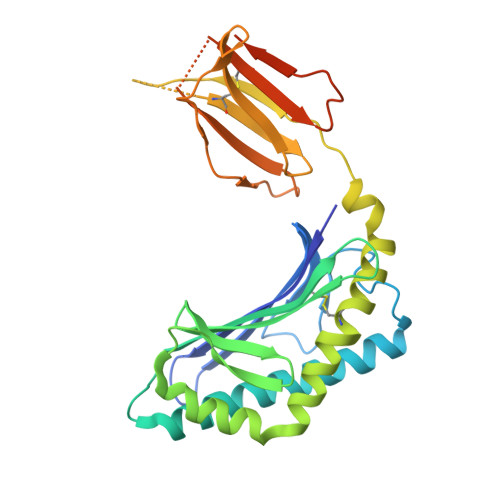
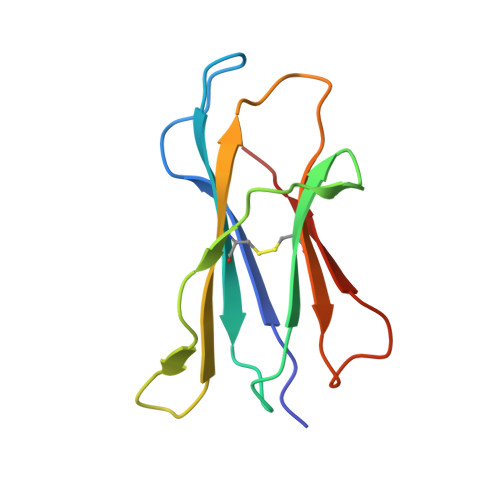
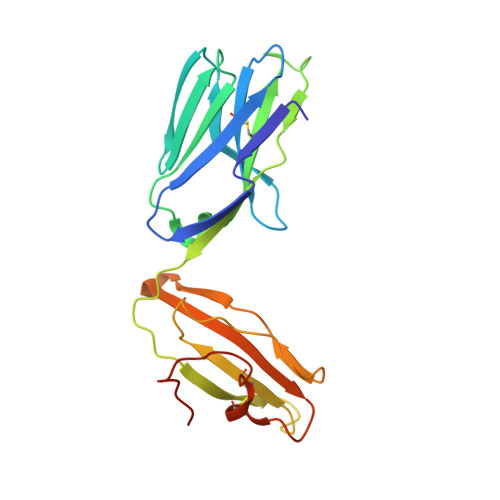
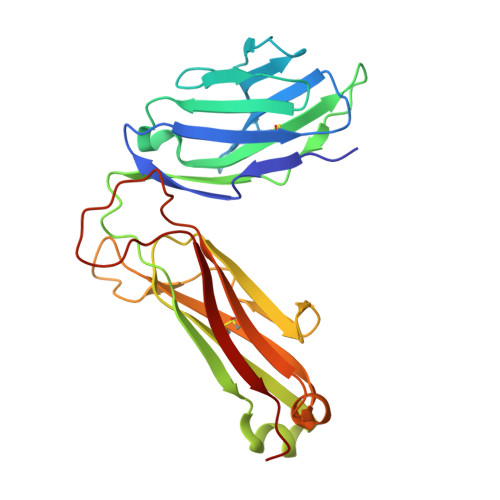
Dual Modifications of alpha-Galactosylceramide Synergize to Promote Activation of Human Invariant Natural Killer T Cells and Stimulate Anti-tumor Immunity.
Chennamadhavuni, D., Saavedra-Avila, N.A., Carreno, L.J., Guberman-Pfeffer, M.J., Arora, P., Yongqing, T., Koay, H.F., Godfrey, D.I., Keshipeddy, S., Richardson, S.K., Sundararaj, S., Lo, J.H., Wen, X., Gascon, J.A., Yuan, W., Rossjohn, J., Le Nours, J., Porcelli, S.A., Howell, A.R.(2018) Cell Chem Biol 25: 571
- PubMed: 29576533
- DOI: https://doi.org/10.1016/j.chembiol.2018.02.009
- Primary Citation of Related Structures:
6BNK, 6BNL - PubMed Abstract:
Glycosylceramides that activate CD1d-restricted invariant natural killer T (iNKT) cells have potential therapeutic applications for augmenting immune responses against cancer and infections. Previous studies using mouse models identified sphinganine variants of α-galactosylceramide as promising iNKT cell activators that stimulate cytokine responses with a strongly proinflammatory bias. However, the activities of sphinganine variants in mice have generally not translated well to studies of human iNKT cell responses. Here, we show that strongly proinflammatory and anti-tumor iNKT cell responses were achieved in mice by a variant of α-galactosylceramide that combines a sphinganine base with a hydrocinnamoyl ester on C6″ of the sugar. Importantly, the activities observed with this variant were largely preserved for human iNKT cell responses. Structural and in silico modeling studies provided a mechanistic basis for these findings and suggested basic principles for capturing useful properties of sphinganine analogs of synthetic iNKT cell activators in the design of immunotherapeutic agents.
Organizational Affiliation:
Department of Chemistry, The University of Connecticut, Storrs, CT 06269-3060, USA.