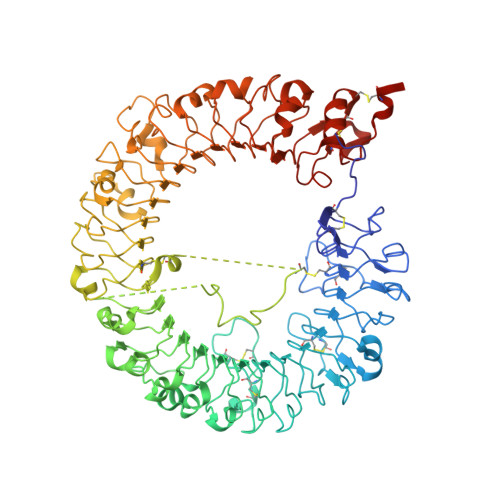
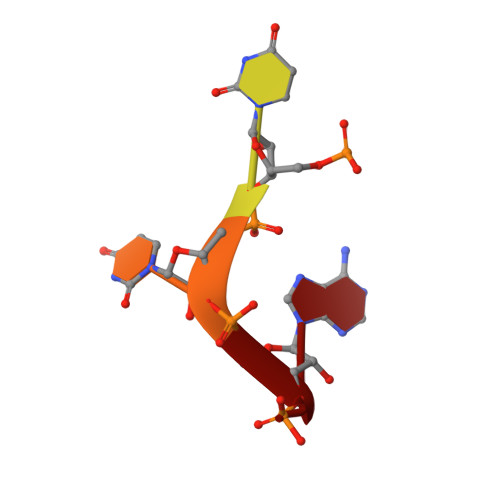
Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Zhang, Z., Ohto, U., Shibata, T., Taoka, M., Yamauchi, Y., Sato, R., Shukla, N.M., David, S.A., Isobe, T., Miyake, K., Shimizu, T.(2018) Cell Rep 25: 3371-3381.e5
- PubMed: 30566863
- DOI: https://doi.org/10.1016/j.celrep.2018.11.081
- Primary Citation of Related Structures:
5ZSA, 5ZSB, 5ZSC, 5ZSD, 5ZSE, 5ZSF, 5ZSG, 5ZSH, 5ZSI, 5ZSJ, 5ZSL, 5ZSM, 5ZSN, 6IF5 - PubMed Abstract:
Toll-like receptor 7 (TLR7) is an innate immune receptor for single-stranded RNA (ssRNA) and has important roles in infectious diseases. We previously reported that TLR7 shows synergistic activation in response to two ligands, guanosine and ssRNA. However, the specific ssRNA sequence preference, detailed recognition mode of TLR7 and its ligand, and molecular determinants of TLR7 and TLR8 selectivity remain unknown. Here, we report on TLR7 from a large-scale crystallographic study combined with a multifaceted approach. We reveal that successive uridine-containing ssRNAs fully or moderately bind TLR7, whereas single uridine-containing ssRNAs have reduced affinities. We also reveal the detailed relationships between the chemical structures of ligands and their binding to TLR7. We demonstrate that an engineered TLR8 mutant alters its responsiveness to TLR7-specific ligands. Finally, we identify guanosine 2',3'-cyclic phosphate (2',3'-cGMP) as a possible endogenous ligand for TLR7 with greater affinity than guanosine. The abundant structural information will facilitate future development of treatments targeting TLR7.
Organizational Affiliation:
Graduate School of Pharmaceutical Sciences, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan.