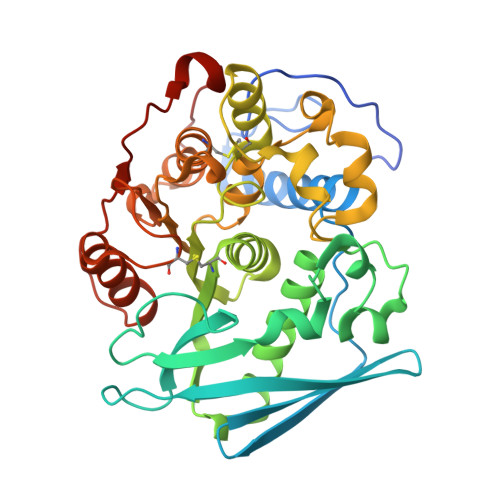
Structure of the catalytic domain of glucuronoyl esterase Cip2 from Hypocrea jecorina.
Pokkuluri, P.R., Duke, N.E., Wood, S.J., Cotta, M.A., Li, X.L., Biely, P., Schiffer, M.(2011) Proteins 79: 2588-2592
- PubMed: 21661060
- DOI: https://doi.org/10.1002/prot.23088
- Primary Citation of Related Structures:
3PIC - PubMed Abstract:
The structure of the catalytic domain of glucuronoyl esterase Cip2 from the fungus H. jecorina was determined at a resolution of 1.9 Å. This is the first structure of the newly established carbohydrate esterase family 15. The structure has revealed the residues Ser278-His411-Glu301 present in a triad arrangement as the active site. Ser278 is present in the novel consensus sequence GCSRXG reported earlier in the members of CE-15 family. The active site is exposed on the surface of the protein which has implications for the ability of the enzyme to hydrolyze ester bonds of large substrates. Efforts are underway to obtain crystals of Cip2_GE complexed with inhibitor and synthetic substrates. The activity of the glucuronoyl esterase could play a significant role in plant biomass degradation as its expected role is to separate the lignin from hemicelluloses by hydrolysis of the ester bond between 4-O-methyl-D-glucuronic acid moieties of glucuronoxylans and aromatic alcohols of lignin.
Organizational Affiliation:
Biosciences Division, Argonne National Laboratory, Argonne, Illinois 60439, USA. [email protected]