Nuclear GTPSCS functions as a lactyl-CoA ligase to promote histone lactylation and glioma progression
Liu, R.L., Ren, X.L., Feng, H.X., Sheng, X.L., Li, L.T., Zhang, Y., Huang, H., Zhao, Y.M.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Succinate--CoA ligase [ADP/GDP-forming] subunit alpha, mitochondrial | 321 | Homo sapiens | Mutation(s): 0 Gene Names: SUCLG1 EC: 6.2.1.4 (PDB Primary Data), 6.2.1.5 (PDB Primary Data) | 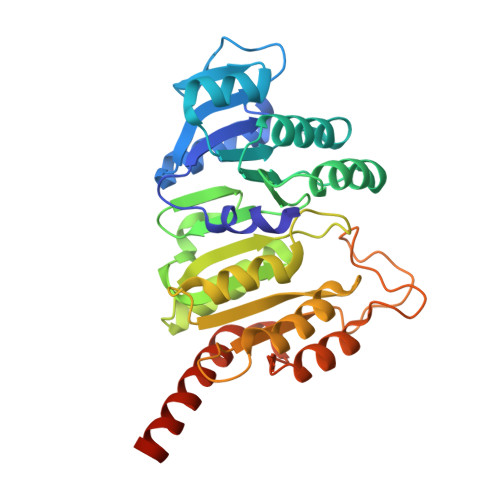 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P53597 (Homo sapiens) Explore P53597 Go to UniProtKB: P53597 | |||||
PHAROS: P53597 GTEx: ENSG00000163541 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P53597 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Succinate--CoA ligase [GDP-forming] subunit beta, mitochondrial | 395 | Homo sapiens | Mutation(s): 0 Gene Names: SUCLG2 EC: 6.2.1.4 | 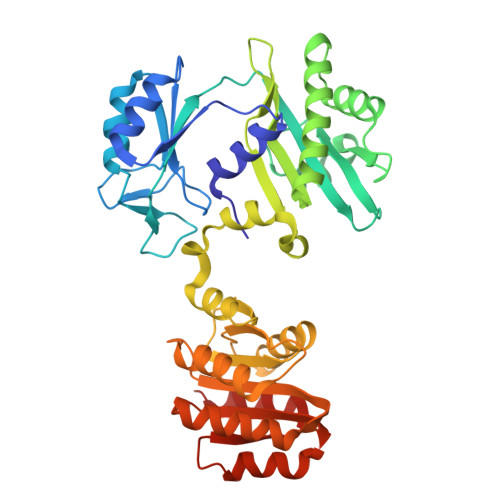 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q96I99 (Homo sapiens) Explore Q96I99 Go to UniProtKB: Q96I99 | |||||
PHAROS: Q96I99 GTEx: ENSG00000172340 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q96I99 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| COA (Subject of Investigation/LOI) Query on COA | C [auth A] | COENZYME A C21 H36 N7 O16 P3 S RGJOEKWQDUBAIZ-IBOSZNHHSA-N |  | ||
| PO4 (Subject of Investigation/LOI) Query on PO4 | D [auth A] | PHOSPHATE ION O4 P NBIIXXVUZAFLBC-UHFFFAOYSA-K |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 56.63 | α = 90 |
| b = 105.7 | β = 90 |
| c = 126.77 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| Aimless | data scaling |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | GM135504 |
| National Institutes of Health/National Cancer Institute (NIH/NCI) | United States | CA251677 |
| National Natural Science Foundation of China (NSFC) | China | 81973164 |