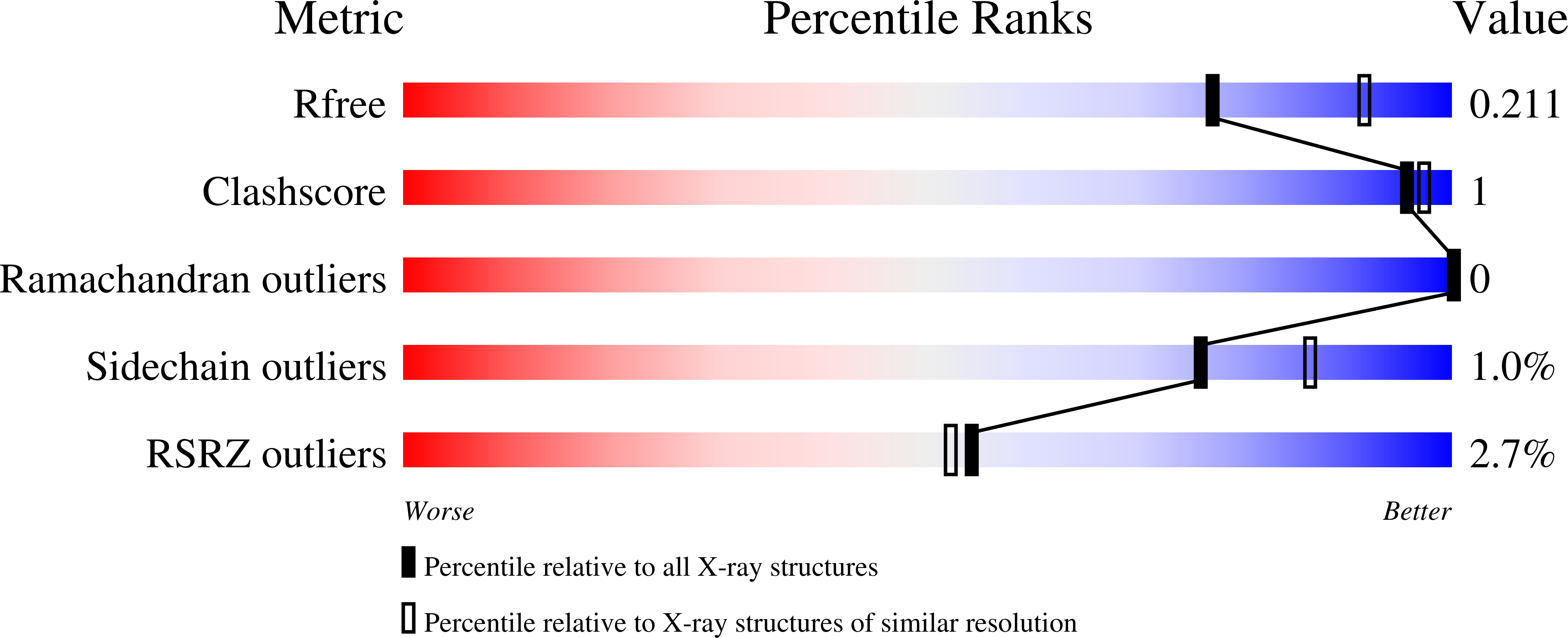
Rational Tuning of Visual Cycle Modulator Pharmacodynamics.
Kiser, P.D., Zhang, J., Badiee, M., Kinoshita, J., Peachey, N.S., Tochtrop, G.P., Palczewski, K.(2017) J Pharmacol Exp Ther 362: 131-145
- PubMed: 28476927
- DOI: https://doi.org/10.1124/jpet.117.240721
- Primary Citation of Related Structures:
5UL5, 5ULG - PubMed Abstract:
Modulators of the visual cycle have been developed for treatment of various retinal disorders. These agents were designed to inhibit retinoid isomerase [retinal pigment epithelium-specific 65 kDa protein (RPE65)], the rate-limiting enzyme of the visual cycle, based on the idea that attenuation of visual pigment regeneration could reduce formation of toxic retinal conjugates. Of these agents, certain ones that contain primary amine groups can also reversibly form retinaldehyde Schiff base adducts, which contributes to their retinal protective activity. Direct inhibition of RPE65 as a therapeutic strategy is complicated by adverse effects resulting from slowed chromophore regeneration, whereas effective retinal sequestration can require high drug doses with potential off-target effects. We hypothesized that the RPE65-emixustat crystal structure could help guide the design of retinaldehyde-sequestering agents with varying degrees of RPE65 inhibitory activity. We found that addition of an isopropyl group to the central phenyl ring of emixustat and related compounds resulted in agents effectively lacking in vitro retinoid isomerase inhibitory activity, whereas substitution of the terminal 6-membered ring with branched moieties capable of stronger RPE65 interaction potentiated inhibition. The isopropyl derivative series produced discernible visual cycle suppression in vivo, albeit much less potently than compounds with a high affinity for the RPE65 active site. These agents were distributed into the retina and formed Schiff base adducts with retinaldehyde. Except for one compound [3-amino-1-(3-isopropyl-5-((2,6,6-trimethylcyclohex-1-en-1-yl)methoxy)phenyl)propan-1-ol (MB-007)], these agents conferred protection against retinal phototoxicity, suggesting that both direct RPE65 inhibition and retinal sequestration are mechanisms of potential therapeutic relevance.
Organizational Affiliation:
Department of Pharmacology, School of Medicine (P.D.K., J.Z., K.P.), Department of Chemistry (M.B., G.P.T.), Case Western Reserve University, Cleveland, Ohio; Research Service, Louis Stokes Cleveland Veterans Affairs Medical Center, Cleveland, Ohio (P.D.K., N.S.P.); Cole Eye Institute, Cleveland Clinic, Cleveland, Ohio (J.K., N.S.P.); and Department of Ophthalmology, Cleveland Clinic Lerner College of Medicine of Case Western Reserve University, Cleveland, Ohio (N.S.P.).