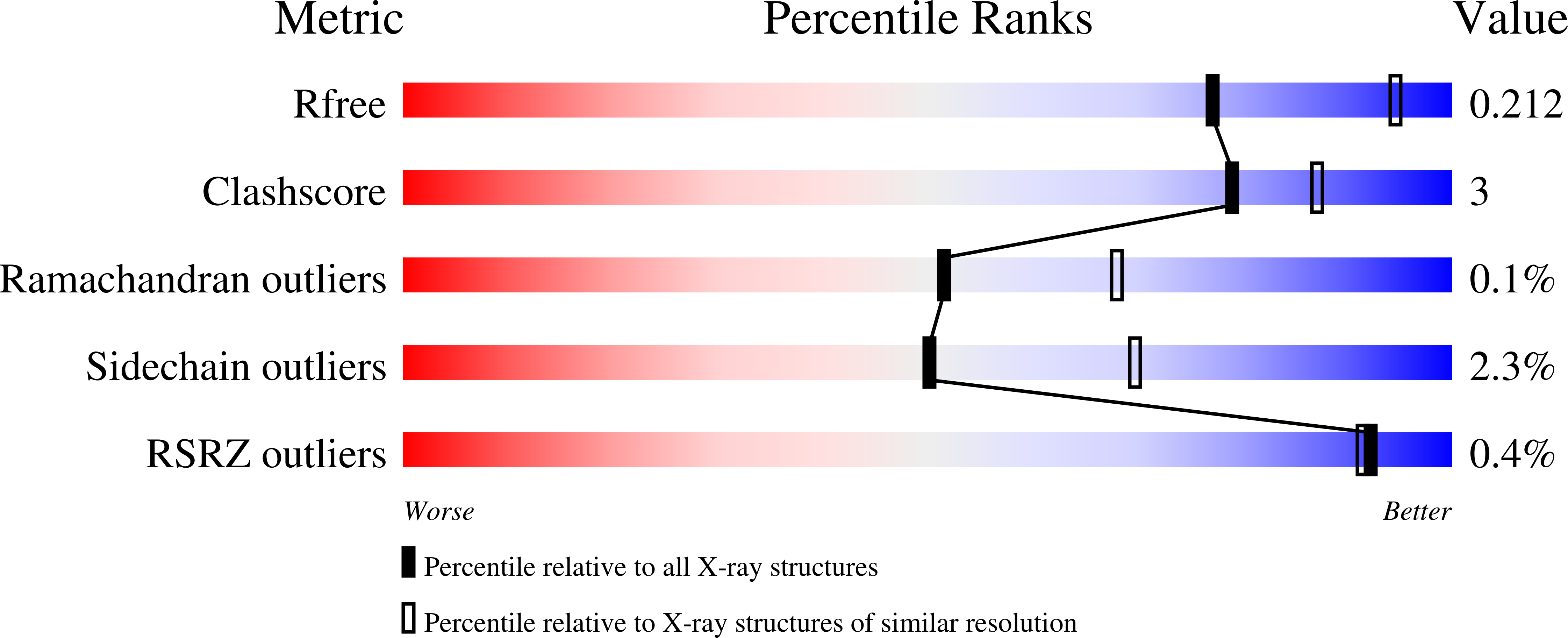
Structural and Functional Influence of the Glycine-Rich Loop G302GGGY on the Catalytic Tyrosine of Histone Deacetylase 8.
Porter, N.J., Christianson, N.H., Decroos, C., Christianson, D.W.(2016) Biochemistry 55: 6718-6729
- PubMed: 27933794
- DOI: https://doi.org/10.1021/acs.biochem.6b01014
- Primary Citation of Related Structures:
5THS, 5THT, 5THU, 5THV - PubMed Abstract:
Histone deacetylase 8 (HDAC8) catalyzes the hydrolysis of acetyl-l-lysine to yield products l-lysine and acetate through a mechanism in which a nucleophilic water molecule is activated by a histidine general base and a catalytic metal ion (Zn 2+ or Fe 2+ ). Acetyl-l-lysine also requires activation by metal coordination and a hydrogen bond with catalytic tyrosine Y306, which also functions in transition state stabilization. Interestingly, Y306 is located in the conserved glycine-rich loop G 302 GGGY. The potential flexibility afforded by the tetraglycine segment may facilitate induced-fit conformational changes in Y306 between "in" and "out" positions, as observed in related deacetylases. To probe the catalytic importance of the glycine-rich loop in HDAC8, we rigidified this loop by preparing the G302A, G303A, G304A, and G305A mutants and measured their steady state kinetics and determined their X-ray crystal structures. Substantial losses of catalytic efficiency are observed (10-500-fold based on k cat /K M ), particularly for G304A HDAC8 and G305A HDAC8. These mutants also exhibit the greatest structural changes for catalytic tyrosine Y306 (1.3-1.7 Å shifts of the phenolic hydroxyl group). Molecular dynamics simulations further indicate that G304 and G305 undergo pronounced structural changes as residue 306 undergoes a transition between "in" and "out" conformations. Thus, the G304A and G305A substitutions likely compromise the position and conformational changes of Y306 required for substrate activation and transition state stabilization. The G302A and G303A substitutions have less severe catalytic consequences, and these substitutions may influence an internal channel through which product acetate is believed to exit.
Organizational Affiliation:
Roy and Diana Vagelos Laboratories, Department of Chemistry, University of Pennsylvania , Philadelphia, Pennsylvania 19104-6323, United States.