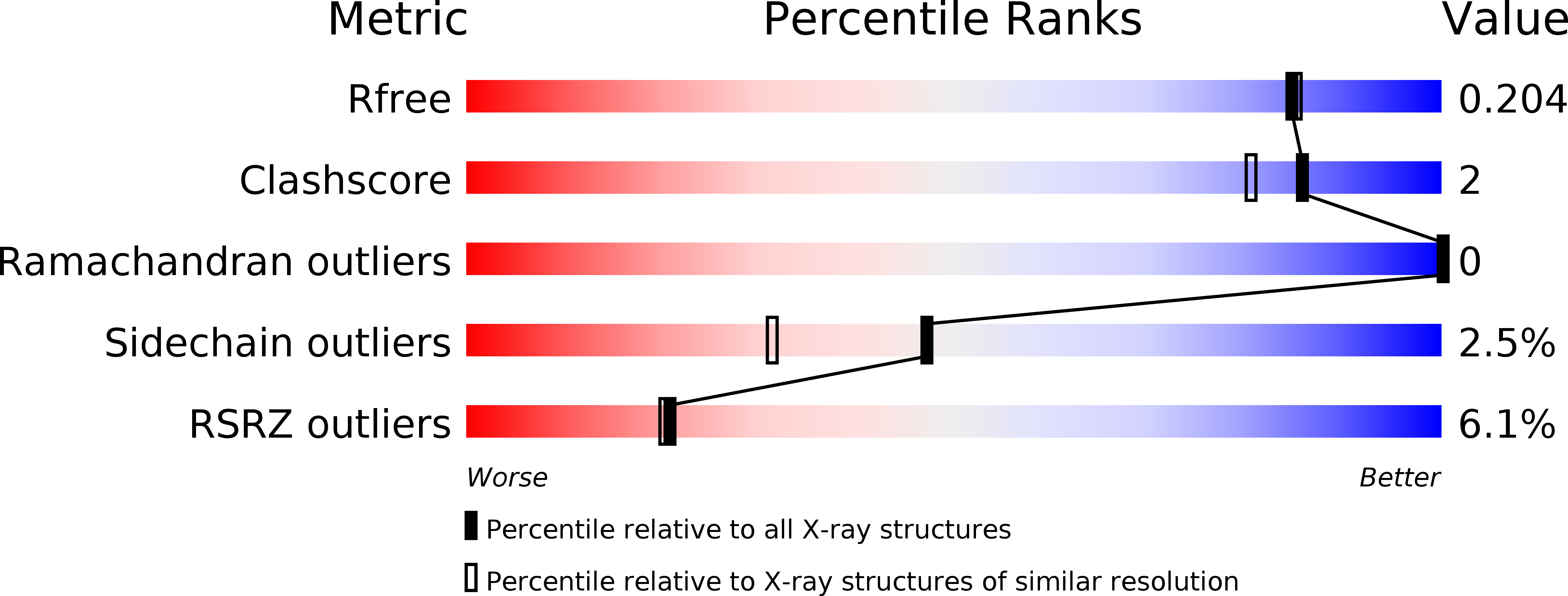
Utilizing Structures of CYP2D6 and BACE1 Complexes To Reduce Risk of Drug-Drug Interactions with a Novel Series of Centrally Efficacious BACE1 Inhibitors.
Brodney, M.A., Beck, E.M., Butler, C.R., Barreiro, G., Johnson, E.F., Riddell, D., Parris, K., Nolan, C.E., Fan, Y., Atchison, K., Gonzales, C., Robshaw, A.E., Doran, S.D., Bundesmann, M.W., Buzon, L., Dutra, J., Henegar, K., LaChapelle, E., Hou, X., Rogers, B.N., Pandit, J., Lira, R., Martinez-Alsina, L., Mikochik, P., Murray, J.C., Ogilvie, K., Price, L., Sakya, S.M., Yu, A., Zhang, Y., O'Neill, B.T.(2015) J Med Chem 58: 3223-3252
- PubMed: 25781223
- DOI: https://doi.org/10.1021/acs.jmedchem.5b00191
- Primary Citation of Related Structures:
4XRY, 4XRZ, 4XXS - PubMed Abstract:
In recent years, the first generation of β-secretase (BACE1) inhibitors advanced into clinical development for the treatment of Alzheimer's disease (AD). However, the alignment of drug-like properties and selectivity remains a major challenge. Herein, we describe the discovery of a novel class of potent, low clearance, CNS penetrant BACE1 inhibitors represented by thioamidine 5. Further profiling suggested that a high fraction of the metabolism (>95%) was due to CYP2D6, increasing the potential risk for victim-based drug-drug interactions (DDI) and variable exposure in the clinic due to the polymorphic nature of this enzyme. To guide future design, we solved crystal structures of CYP2D6 complexes with substrate 5 and its corresponding metabolic product pyrazole 6, which provided insight into the binding mode and movements between substrate/inhibitor complexes. Guided by the BACE1 and CYP2D6 crystal structures, we designed and synthesized analogues with reduced risk for DDI, central efficacy, and improved hERG therapeutic margins.
Organizational Affiliation:
#The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, California 92024, United States.